1. Alzheimer’s disease (AD): AD is the most common cause of dementia, manifested as memory loss and other cognitive impairments that interfere with daily life. Converging evidence supports the notion that AD is the result of selective vulnerability of specific neural networks. Yet, our molecular understanding of selectively vulnerable neural networks is very limited. Only by understanding the molecular phenotype of these cellular networks can we devise strategies to protect these networks from degeneration.
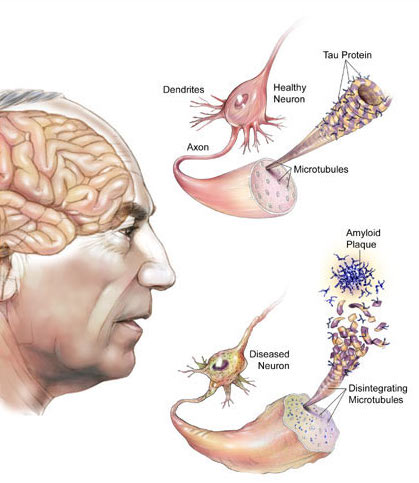
In collaboration with Dr. Edward Lee, we are studying cellular transcriptomic variations from human brains with the hope of providing a detailed molecular phenotype of vulnerable cellular networks as a foundation for developing therapeutic strategies for AD.
2. Age-related macular degeneration (AMD): AMD affects over 10 million Americans, twice the number affected by Alzheimer’s disease and equal to the total of all cancer patients combined. AMD is multifactorial, driven by both genetic and environmental factors. Despite being the first disease successfully studied by GWAS, it is still unknown how the GWAS findings can be translated into therapeutic targets. There is an urgent need to collect and analyze retinal cells from human eyes to advance our understanding of AMD.
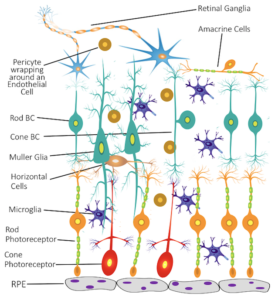
In collaboration with Dr. Dwight Stambolian and collaborators at the University of Alabama, we are studying single-cell transcriptomic variations in human eyes. We hope to provide a detailed characterization of human retinal cell atlas and novel insights into cell-type-specific functions that can power precision therapeutic targeting of AMD.
3. Cardiometabolic diseases (CMD): Although monocytes and macrophages play central roles in atherosclerosis and related CMD, we have yet to realize monocyte targeted therapeutics from human CMD. In collaboration with Dr. Muredach Reilly at Columbia University, we are performing single-cell RNA-seq profiling of human monocytes coupled to innovative population genetics and functional genomics to define the role of human monocyte subpopulations inhuman monocyte-macrophage biology and CMD.
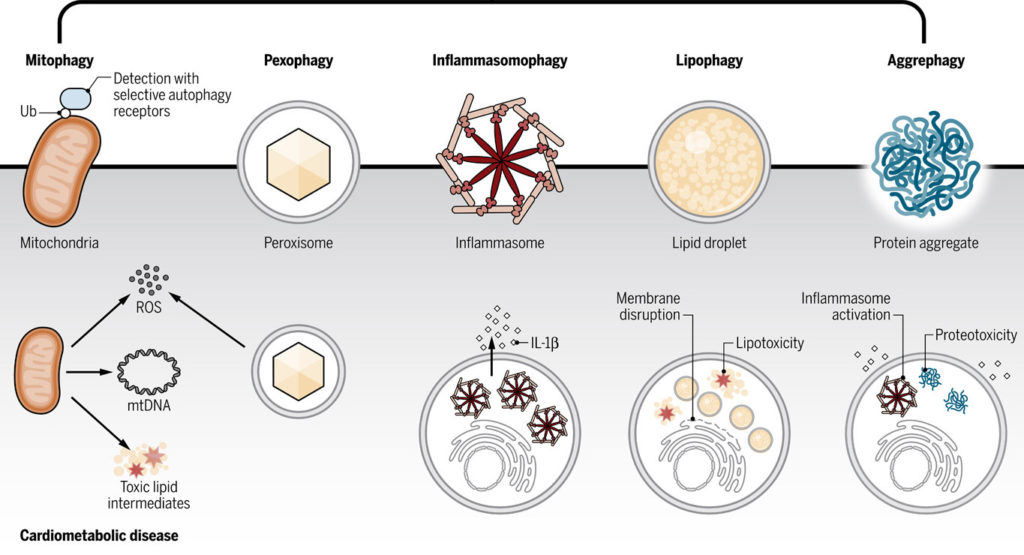
We hope to address a major knowledge gap in human monocyte biology by defining the landscape of human monocyte subpopulations, their regulatory features, genetic association with CMD, and functional genomics in human model systems.
4. Type 1 Diabetes (T1D): T1D is the primary form of diabetes affecting children and adolescents, with an alarming increase in incidence of 3%-5% per year. The cellular and molecular events underlying T1D initiation and progression in humans are, so far, poorly defined. Although T1D is thought to be a T cell mediated disease, it is well known that the delicate interplay of cell types of the innate and adaptive immune system determines disease outcome. It is yet unclear the precise role that each cell type plays, especially in the early disease stages.

In this collaboration with Dr. Ali Naji and the NIDDK Human Pancreas and Analysis Program (HPAP) at the University of Pennsylvania, we apply single cell RNA sequencing to profile human pancreatic islets and surrounding lymph nodes during homeostasis and T1D autoimmune attack.
5. Cancer: Cancer is characterized by inter-cellular diversity, where clonal subpopulations of tumor cells are intermixed with stromal cells and infiltrating immune cells. Each subclone of the tumor carries its own set of genetic variants and transcriptional signatures, whereinter-population DNA variation is compounded with cell-to-cell RNA expression stochasticity. Characterizing this genomic and transcriptomic diversity require analyses at the resolution of individual cells and contiguous genome molecules.

In this collaboration with Stanford University, Perelman School of Medicine at University of Pennsylvania, and Children’s Hospital of Philadelphia, we develop computational methods to jointly analyze transcriptomic and genetic intratumor diversity and apply these to the study of a variety of cancer types.
